Skin is now composed of a human part and a microbial part which is the more variable part. Up to now a lot of information could be obtained at the diversity, richness and taxonomic levels of microbiotas. Clinical trials are able to show effects which are documented by these descriptive levels but few explanation of effects are clearly described. Either way, it becomes essential to understand the mechanisms of action and the interaction of the human part with the microbial part to be able to master the safety and efficacy on the skin of drugs and cosmetics. This is possible with omics techniques, which however have different capacities and performances to answer these questions. Furthermore, it is now clear with the science of the microbiome that interactions such as the skin-gut axis are of paramount importance, which means that any knowledge of the microbiota can be beneficial for the skin microbiota understanding. Gilbert Skorski, CEO of Phylogene, explains.
Technical landscape
PCR was discovered around 1985 and allowed to detect specifically DNA strands. Since the years 1990, DNA sequencing techniques opened the door to untargeted approaches of genes. Now NGS allows sequencing a genome at low cost.
Since 1945, mass spectrometry allowed detection of small chemical molecules. For 15 years now, mass spectrometry evolution allows to analyze and quantify proteins. The proteogenomics approach through the genes knowledge and with LC-MS/MS and databases evolutions allows identifying and quantifying proteins in an untargeted mode. Improvements in the mass accuracy, resolution, and sensitivity of MS instruments are enabling the rapid and reliable detection, identification, and quantification of proteins in complex mixtures.
These techniques are opening the door to improved methods for discovering disease-specific biomarkers with the potential to support early disease detection and even individualized therapies.
Fig 1: Techniques are operational at different steps of the systems biology and provide different levels of information which are targeted specifically on analytes or only assigns a signal to an analyte through databases.
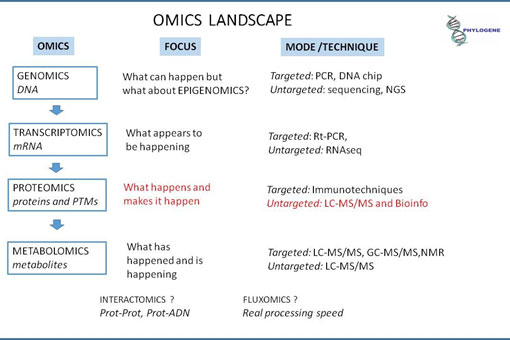
In an untargeted way (shotgun NGS) and quantitatively, a gene gives the information that the translated protein can structure or work in the cell, but we now know …